Research Goals
Cell Biology Group aims to bring about new ICT paradigms based on information processing and communication technologies in living cells. Our study focuses on the following three areas...
Analysis and application of cellular information systems
The genetic information system in cells is an excellent product of biological evolution for 3.5 billion years. We are clarifying the regulatory principles and molecular mechanisms of the genetic information system in order to create new ICT based on the cellular systems.
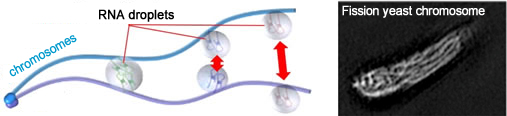
We have found that RNA–protein complexes assembled at specific chromosomal loci mediate recognition and subsequent pairing of homologous chromosomes through liquid-phase separation. The discovery reveals one of the mechanisms for communication through bio macromolecules.
Ding et al., (2019) Nat Commun, 10, 5598
Ding et al., (2012) Science, 336: 732-736.
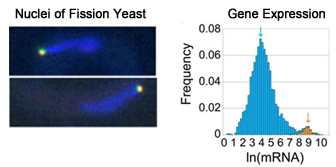
Using genome-wide gene expression analyses, we have identified factors responsible for the chromosomal arrangement in the nucleus of distinct developmental stages and unraveled the mechanism for the specific chromosomal arrangement. Now we are working on to understand the mechanism regulating global gene expression over the entire chromosomes.
Chikashige et al., (2015) Sci Rep., 5: 15617
Chikashige et al., (2009) J. Cell Biol. 187: 413-427
Chikashige, et al., (2006). Cell 125, 59-69.
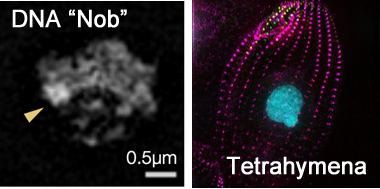
Using super-resolution microscopy, we have discovered a novel DNA region and have also been working on to understand the mechanism that forms cellular cortex structures and cellular morphology. The mechanism that forms morphology of single cells is difficult to accessed by the classical approaches like genetics, molecular biology and biochemistry, and thus is as yet unknown. If we could understand this mechanism, then we would be able to control cellular morphology at will or able to exploit self-organized, efficient biomaterials. Unicellular organism Tetrahymena has a functional cellular shape that is known to be formed as a result of patterned arrangement of cellular organelles according to some geometric mathematical rules. Although classical studies were unable to understand this mechanism, we are trying to understand it using new technologies including super-resolution microscopy.
Lowe et al., (2021) eLife 10:e65369
Murawska et al., (2019) Mol Cell, 77:501-513.
Tashiro et al. (2016) Nat Commun, 7: 10393
Matsuda et al., (2015) Nat Commun, Jul 24; 6:7753.
Development of bio-imaging technology
Development of the fluorescence imaging technology that visualizes the behavior of target molecules in living cells is an important part of our study to monitor the information flow inside cells. We have developed microscopy methods including live-imaging methods with reduced cytotoxicity, and chromatic correction and artifact reduction in super-resolution microscopy. We also released our software for researchers in this field and held a yearly workshop for microscopy over many years. Fluorescence microscopy is essential for us to exploit the cellular ICT.

Postdoctoral position (1) available!
Postdoctoral position (2) available!
Tahara et al., (2021) Applied Optics, 60:A260-A267
Ashida et al., (2020) J. Biomed. Opt, 25:123707
Matsuda et al., (2020) J Vis Exp, (160), e60800
Matsuda et al., (2018) Sci Rep, 8:7583
Kraus et al., (2017) Nat Protoc, 12:1011-1028
Demmerle et al, (2017) Nat Protoc, 12:988-1010
Construction of intracellular structures and control of cellular function
We are developing the technology to construct artificial structures in living cells that will allow us to control cellular function. Construction of functioning artificial organelles in cells will enable us to create various artificial cells that can act as sensors for specific substances, perform drugs screening, and generate various useful materials.
Bilir, et al. (2019) Genes to cells 24, 338-353.
Kobayashi, et al. (2015) Proc. Natl Acad. Sci. 112, 7027-7032.
Kobayashi, et al. (2010) Autophagy 6, 36-45.